Mike and Stephanie’s work regarding PDAC tumor model is published at the Journal “Small.” This paper aims to create an in vitro tumor model reconstituting intratumoral heterogeneity of human pancreatic cancer. This work is based on collaboration with Professor Konieczny’s group in the Department of Biological Science at Purdue.
A Biomimetic Tumor Model of Heterogeneous Invasion in Pancreatic Ductal Adenocarcinoma
Michael J. Bradney, Stephanie M. Venis, Yi Yang, Stephen F. Konieczny, and Bumsoo Han
Abstract
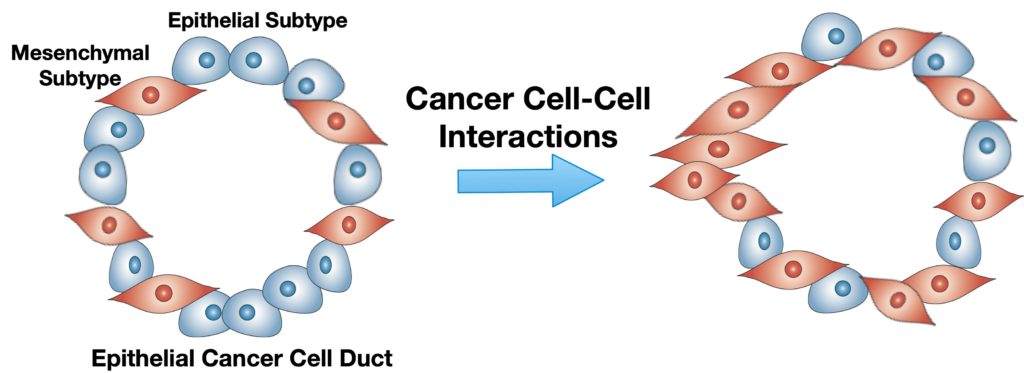
Pancreatic ductal adenocarcinoma (PDAC) is a complex, heterogeneous, and genetically unstable disease. Its tumor microenvironment (TME) is complicated by heterogeneous cancer cell populations and strong desmoplastic stroma. This complex and heterogeneous environment makes it challenging to discover and validate unique therapeutic targets. Reliable and relevant in vitro PDAC tumor models can significantly advance the understanding of the PDAC TME and may enable the discovery and validation of novel drug targets. In this study, an engineered tumor model is developed to mimic the PDAC TME. This biomimetic model, named ductal tumor‐microenvironment‐on‐chip (dT‐MOC), permits analysis and experimentation on the epithelial–mesenchymal transition (EMT) and local invasion with intratumoral heterogeneity. This dT‐MOC is a microfluidic platform where a duct of murine genetically engineered pancreatic cancer cells is embedded within a collagen matrix. The cancer cells used carry two of the three mutations of KRAS, CDKN2A, and TP53, which are key driver mutations of human PDAC. The intratumoral heterogeneity is mimicked by co‐culturing these cancer cells. Using the dT‐MOC model, heterogeneous invasion characteristics, and response to transforming growth factor‐beta1 are studied. A mechanism of EMT and local invasion caused by the interaction between heterogeneous cancer cell populations is proposed.